Architecture
SoyKB has four modules including the MySQL database module at the back end that incorporates and integrates all the soybean genomics and omics data from various experiments. It is designed to contain information on 6 different entities namely genes/proteins, miRNAs/sRNAs, metabolites, SNPs, Plant Introduction (PI) lines, and traits. The other three front-end modules are web interface, genome browser and data integration.
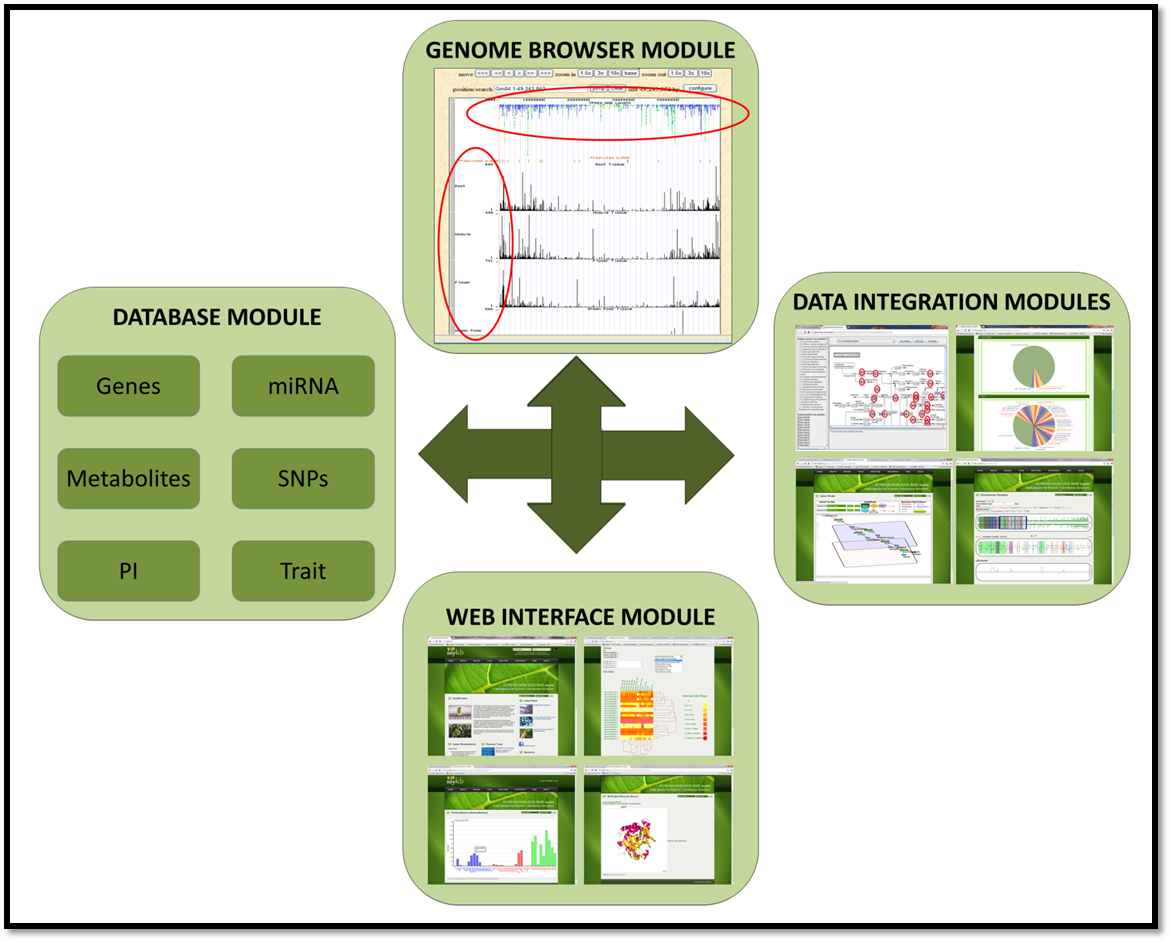

1 Database Module
The database module includes a mySQL database to incorporate and integrate all the soybean raw data from various types of experiments. It is designed to contain information on 6 different entities namely genes/proteins, miRNAs/sRNAs, metabolites, SNPs, Plant Introduction (PI) lines, and traits. The experimental data in the database are generated by the labs of Gary Stacey and Henry Nguyen, together with data from public sources.
2 Web Interface
This module is designed using PHP to provide access to the stored information through a web interface, where biologists and soybean producers can search and retrieve information about genes of interest, access the experimental observations from different experimental conditions and integrate information with the pathways. Special attention has been paid to the security and permissions of the site controlled by a mandatory, user sign-in requirement.
3 Genome Browser
All the data in SoyKB has been deposited into the genome browser, which is set up locally for soybean utilizing the architecture provided by UCSC. The module allows users to visualize the genomic information on entire chromosome and help understand the overall picture. The browser allows users to zoom in and out to focus on regions of their interest and gives the users the flexibility to load and/or hide the experimental tracks they wish to visualize.
4 Data Integration
This module is targeted towards integrating the data from various experimental conditions and portraying the information in various integrative tools to highlight the expressed genes/proteins and metabolites based on the selected microarray, proteomics, transcriptomics and metabolomics data respectively. Many tools are available including In Silico Breeding Program, differential expression analysis suite of tools and pathway viewers.
Latest News
 SoyKB: a powerful tool at the junction of plant biology and computer science
SoyKB: a powerful tool at the junction of plant biology and computer science
 SoyKB: a powerful tool at the junction of plant biology and computer science
SoyKB: a powerful tool at the junction of plant biology and computer science
 SoyKB Facebook
SoyKB Facebook
SoyKB workshop @ Soy2014
Molecular and Cellular Biology of the Soybean 15th Biennial Conference
Molecular and Cellular Biology of the Soybean 15th Biennial Conference






