- Home
- Search
- Browse
- Tools
Affymetrix Probe ID Mapper Pathway Viewer MotifSampler w/ WebLOGO Blast Multiple Sequence Alignment Phylogeny Chromosome Visualizer Protein BioViewer Heatmap and Hierarchical Clustering ScatterPlot SNPViz Targeted Sequence Selection Distance Matrix Mutant Finder Enrichment Analysis SoyCSN AccuTool Soybean Protein Sequence Logos Soybean Allele Catalog Tool Soybean Genomic Variations Explorer Soybean MADis Tool
- SoyHUB
- Data Files
- Analytics
- Information
- About
NGS RESEQUENCING BROWSER
Introduction
With the advances in next generation sequencing (NGS) technology and significant reduction in sequencing costs it is now possible to sequence large sets of crop germplasm and generate whole genome scale structural variations and genotypic data. In depth informatics analysis of the genotypic data can provide better understanding of the links with the observed phenotypic changes. This approach can be used to further understand and study different traits for the improvement of crops by design.
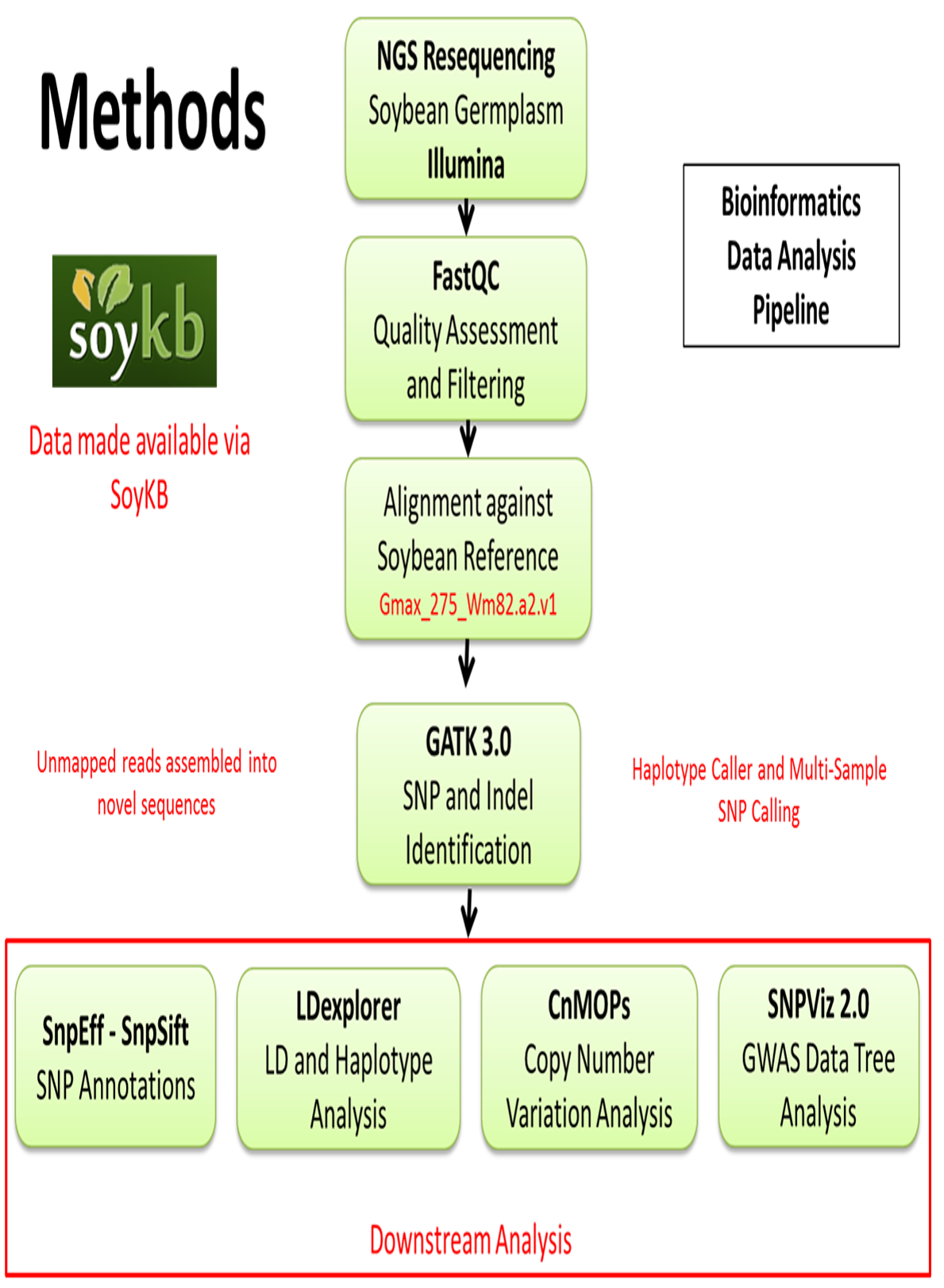
Bioinformatics Analysis Pipeline
We have built a bioinformatics analysis pipeline for identification of SNPs and insertion, deletions using GATK3.0 software against soybean reference genome Gmax_275_Wm82.a2.v1. Downstream analysis is conducted for copy number variations (CNV) using CnMOPs and SNP annotations generated using SnpEff and SnpSift. Data is also analyzed using LDexplorer and in-house built tool SNPViz.
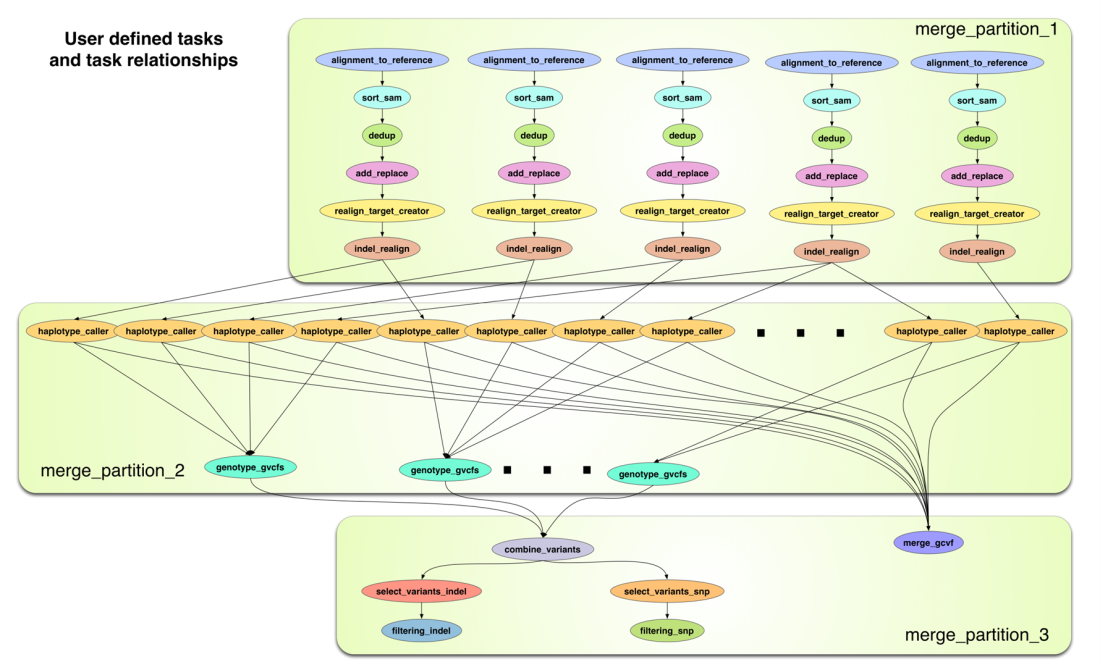
Analysis is conducted using XSEDE as the computing infrastructure, iPlant as the data and cloud infrastructure, and the Pegasus workflow systems to control and coordinate the data management and computational tasks. Each workflow gets mapped into three pegasus-mpi-cluster jobs; one with BWA going to the normal compute nodes, one for large memory Picard and GATK tasks mapped to the large memory nodes available on Stampede, and the last one lower memory GATK tasks. It outlines best practices for efficient utilization of distinct and unique Cyberinfrastructure (CI) resources available through multiple providers, with emphasis on creating extensible and scalable workflows that can be easily modified and deployed.
Data Generation Phases
We have generated resequencing data for several soybean lines from various projects using Illumina paired end sequencing technology. Details of soybean lines and traits selected are as follows:
Phase I: MSMC (15X)
Phase II: USB (40X & 15X)
USB (15X) (planned in future)
Data Access
Data is stored on iPlant Data Store cloud computing resources with controlled access. Data is available for access at multiple tiers including:
* Note: This tool loads results files directly from iPlant data store. Some files may be temporarily inaccessible during the time of iPlant maintenance. Please re-access the link after the maintenance is complete or contact SoyKB team for any questions.
MSMC
USB 40X
USB 15X
* Note: This tool loads results files directly from iPlant data store. Some files may be temporarily inaccessible during the time of iPlant maintenance. Please re-access the link after the maintenance is complete or contact SoyKB team for any questions.
MSMC
USB 40X
USB 15X
* Note: This tool loads results files directly from iPlant data store. Some files may be temporarily inaccessible during the time of iPlant maintenance. Please re-access the link after the maintenance is complete or contact SoyKB team for any questions.
MSMC
USB 40X
USB 15X
* Note: This tool loads results files directly from iPlant data store. Some files may be temporarily inaccessible during the time of iPlant maintenance. Please re-access the link after the maintenance is complete or contact SoyKB team for any questions.
MSMC
USB 40X
USB 15X
* Note: This tool loads results files directly from iPlant data store. Some files may be temporarily inaccessible during the time of iPlant maintenance. Please re-access the link after the maintenance is complete or contact SoyKB team for any questions.
MSMC
USB 40X
USB 15X
* Note: This tool loads results files directly from iPlant data store. Some files may be temporarily inaccessible during the time of iPlant maintenance. Please re-access the link after the maintenance is complete or contact SoyKB team for any questions.
MSMC
USB 40X
USB 15X
* Note: This tool loads results files directly from iPlant data store. Some files may be temporarily inaccessible during the time of iPlant maintenance. Please re-access the link after the maintenance is complete or contact SoyKB team for any questions.
MSMC
SNP
Summary Report
Annotation
Choose Position Range:
Chromosome: Start Position End Position
Input Gene ID(Example: Glyma.01G001000, Glyma.16G175100):
Gene ID
Chromosome: Start Position End Position
Input Gene ID(Example: Glyma.01G001000, Glyma.16G175100):
Gene ID
INDEL
USB 40X
USB 15X
Coming Soon
Coming Soon