SoyHUB
Welcome to SoyHUB
A hub for soybean applied genomics predictions based on curated panels of diverse soybean resequenced accessions (Soy775, Soy1066, and Soy2939).
Explore Variations
Allele Catalog Tool- Visualize alleles, functional effects, amino acid changes in genes - Lookup accessions within selected allele groupings - Map alleles to phenotype to visualize allele and phenotype distributions |
Genomic Variation Explorer- Explore variations in transcription factor binding sites within promoter regions - Explore copy number variations and link copy number variations to genes - Map copy number variations to phenotype to visualize copy number variation and phenotype distributions |
Multiple Allele Discovery Tool- Multiple allele discovery to understand combinative mutations - Perform multiple allele position combinative calculations |
Protein Sequence Logos- Generates sequence logos for protein based on multiple organisms |
Phenotype Distribution Tool- Explore genes that are highly associated with a selected phenotype - Connect genes to Allele Catalog Tool to highlight causative variant positions |
Paralog Allele Catalog Tool- Explore paralogs and connect to Allele Catalog visualization |
Genome Explorer- An interactive tool designed to visualize and analyze the genomic data of soybeans for research and breeding applications |
Genotyping Tool- A platform that enables efficient genotyping and analysis of genetic variations in soybean for improved breeding and research |
Predict New Causal Mutations
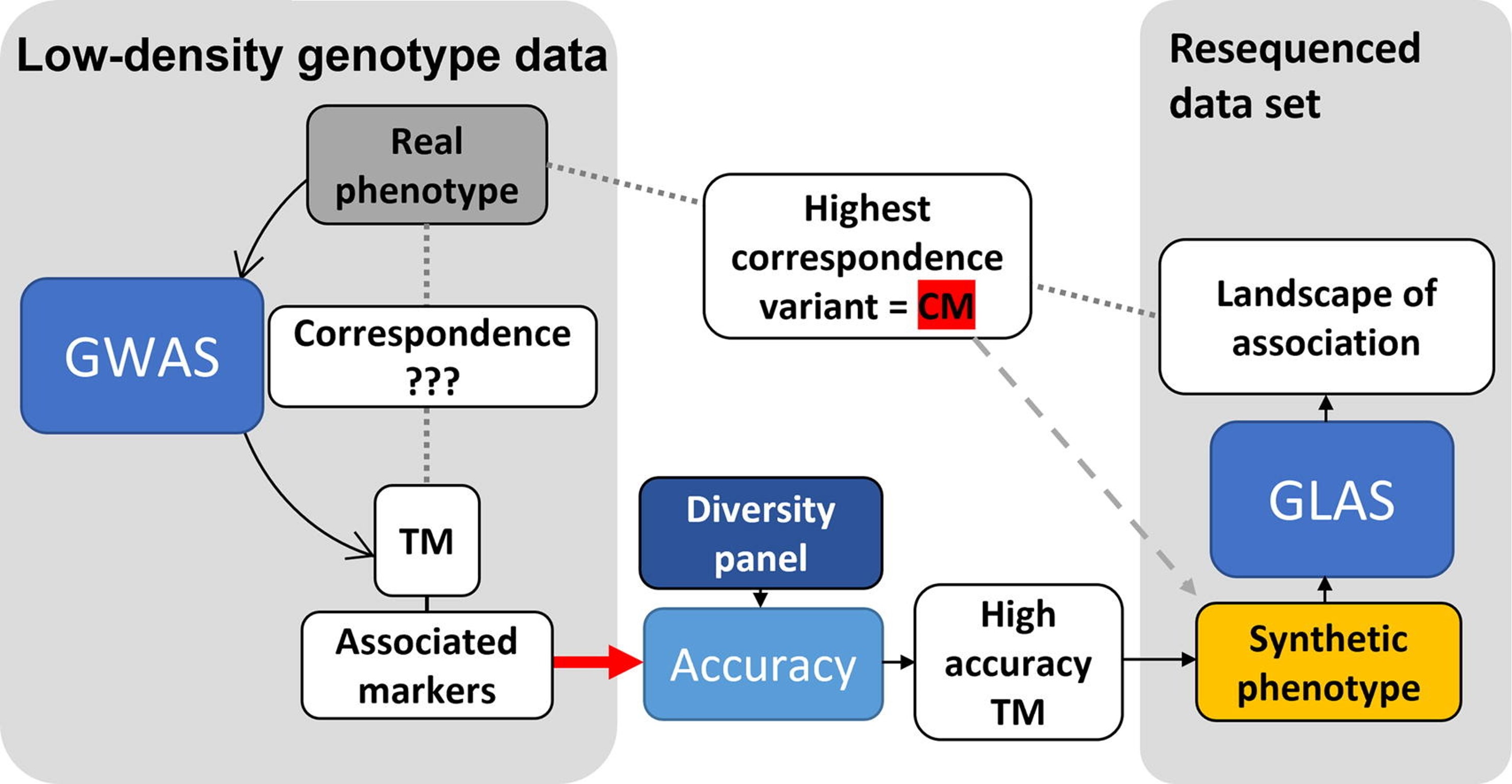
AccuTool- Use GWAS results for prediction - Calculate Accuracy for your markers or candidate causative mutations (CM) based on Soy775 35.7M variant positions |
SNPViz- Check genomic context of your variant positions in empowered haplotype viewer on various resequenced data sets |
Reference Interassembly Gene Browser- Search between reference genotypes, genome assemblies, or annotation versions |
Survey for SoyHUB
We would love to hear from you about your experience with SoyHUB and our genomic analysis tools. Your opinion is valuable to us as it will help us improve our existing tools and develop new features. The survey should only take you a few minutes.
Your responses will be completely anonymous unless you leave your email address to get in touch with us. We appreciate your input!
Click here for Feedback FormReferences
Allele Catalog
[1] Chan YO, Dietz N, Zeng S, Wang J, Flint-Garcia S, Salazar-Vidal MN, Škrabišová M, Bilyeu K, Joshi T: The Allele Catalog Tool: a web-based interactive tool for allele discovery and analysis. BMC Genomics 2023, 24(1):107.
GenVarX
[1] Chan YO, Biova J, Mahmood A, Dietz N, Bilyeu K, Škrabišová M, Joshi T: Genomic Variations Explorer (GenVarX): A Toolset for Annotating Promoter and CNV Regions Using Genotypic and Phenotypic Differences. Frontiers in Genetics 2023.
AccuTool
[1] Biová J, Dietz N, Chan YO, Joshi T, Bilyeu K, Škrabišová M: AccuCalc: A Python Package for Accuracy Calculation in GWAS. Genes 2023, 14(1):123.
[2] Škrabišová M, Dietz N, Zeng S, Chan YO, Wang J, Liu Y, Biová J, Joshi T, Bilyeu KD: A novel Synthetic phenotype association study approach reveals the landscape of association for genomic variants and phenotypes. Journal of Advanced Research 2022, 42:117-133.
MADis
[1] Biová J, Kaňovská I, Chan YO, Immadi MS, Joshi T, Bilyeu K, Škrabišová M. Natural and artificial selection of multiple alleles revealed through genomic analyses. Front Genet. 2024 Jan 8;14:1320652. doi: 10.3389/fgene.2023.1320652. PMID: 38259621; PMCID: PMC10801239.
SNPViz
[1] Zeng S, Škrabišová M, Lyu Z, Chan YO, Dietz N, Bilyeu K, Joshi T: Application of SNPViz v2.0 using next-generation sequencing data sets in the discovery of potential causative mutations in candidate genes associated with phenotypes. International Journal of Data Mining and Bioinformatics 2021, 25(1-2):65-85.
[2] Zeng S, Škrabišová M, Lyu Z, Chan YO, Bilyeu K, Joshi T: SNPViz v2.0: A web-based tool for enhanced haplotype analysis using large scale resequencing datasets and discovery of phenotypes causative gene using allelic variations. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM): 16-19 Dec. 2020; 2020: 1408-1415.
[3] Langewisch T, Zhang H, Vincent R, Joshi T, Xu D, Bilyeu K: Major soybean maturity gene haplotypes revealed by SNPViz analysis of 72 sequenced soybean genomes. PLoS One 2014, 9(4):e94150.